|
Dhruva Abhijit Rajwade I will be joining Prof. Marianna Rapsomaniki's AI/ML For Biomedicine group in CHUV, Lausanne as a Junior Research Scientist, working on uncertainty estimation and generative modeling for biology. I graduated in May'25 as an Integrated Master's student at IIT Kharagpur in India, where I Majored in Biotechnology and Biochemical Engineering and worked in Computational Biology, Computer Vision and AI4Science. In my undergrad journey, I have worked with Prof. Koel Chaudhary and Prof. Soumya De at IIT Kharagpur. In the summer of 2023, I got an amazing opportunity to work with Prof. Brian Ingalls at the University Of Waterloo. I worked at Caltech in the summer of '24 with Prof. Anima Anandkumar and Dr. Shengchao Liu on understanding Protein-DNA interactions using Cross-Attention and language modelling. Currently, I am wrapping up my Master's thesis on using discrete diffusion for controllable generation of DNA-binding protein sequences, and am excited to see where this project goes. I am a recipient of the Caltech SURF fellowship (2024), the MITACS Globalink Research Internship (2023), and have also been selected for the EPFL E3 scholarship (2024) and the ThinkSwiss Research scholarship (2023). |


|
ResearchI'm primarily interested in Deep Learning for Protein Design, Gene regulation and Biological systems, Mathematical modelling of biological networks and dynamics, and AI for science. Most of my work has involved applying robust learning methods to Biological problems in an interpretable manner. Apart from Biology, I am keenly interested in and have also worked in Deep Learning for Vision (specifically SSL, secure ML and Generative modelling), as well as Causal Inference and Graph Learning. More recently, I have been interested in Fourier Neural Operators, Geometric Deep Learning, State-space models and their applications in Biology. |
News |
| [May 2025] | Some early work from my Master's thesis was accepted as a poster at the AI Bio X conference at Sanger, Cambridgeshire. See you in the UK! |
| [Dec 2024] | Our paper on backdoor and adversarial attacks targetting SSL was accepted at ICASSP 2025. See you at Hyderabad! |
| [Nov 2024] | Our work on understanding Protein-DNA interactions using Protein and Genomics Foundation models was accepted at the MLSB, FM4Science and AIDrugX workshops, NeurIPS 2024. |
Current Projects |
|
Discrete Diffusion For Tunable DNA-binding Protein
Design
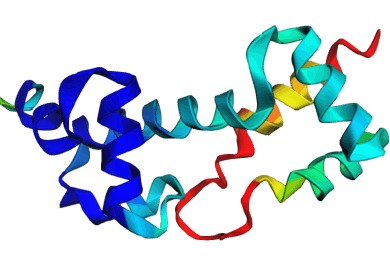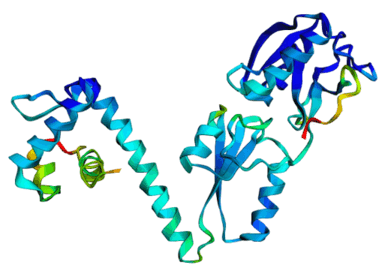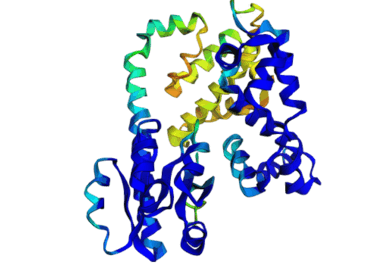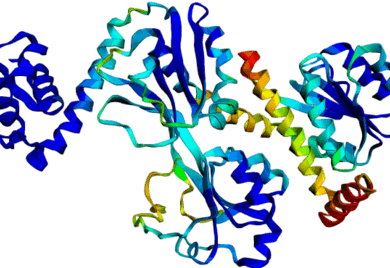
[Slides] Using Discrete Diffusion to learn the distribution of DNA-binding protein sequences. Using our work on Seq2Contact to guide the diffusion process to generate proteins with high affinity for specified DNA targets. Working on optimizing CRISPR-Cas protein design, stitching together different functional domains by inpainting a DNA-binding domain in between, generating DNA sequences for binding to a target protein by inverting the Seq2Contact model, and engineering protein-DNA interactions. This project is in collaboration with Prof. Riddhiman Dhar at IIT Kharagpur. Image shows ESMFold folded sequences sampled by our trained discrete diffusion model, with as less as 25% sequence similarity to the training set and the presence of different DNA-binding domains (through PFam scans). The protein sequences were not cherry-picked, and the structures are coloured by confidence (blue: High, red: Low). |
|
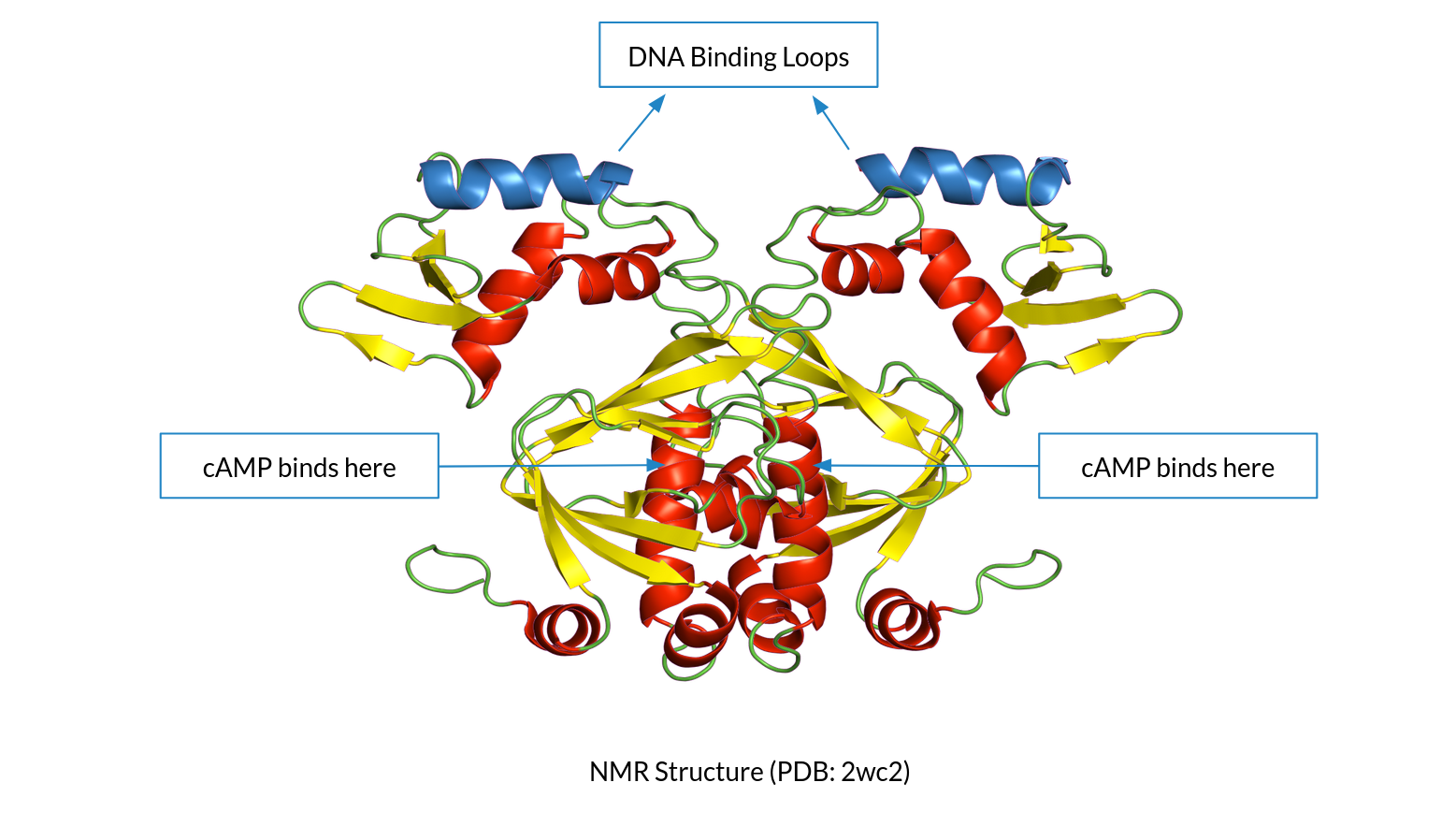
Finding Allosteric Networks in the CAP-cAMP system using
Deep Learning and Molecular Dynamics
[Slides] Using MD simulations and trajectory analysis to infer Allosteric networks in the CAP-cAMP system (where the cAMP ligand binds to CAP protein, leading to allosteric orientation changes and finally, transcription). Current progress includes uncovering of three prospective networks using AlloReverse. Future plans include using State-space models to learn the dynamics of protein-ligand interactions, and finding a generalizable method for allosteric discovery. This work is in collaboration with Prof. Soumya De at IIT Kharagpur. |
Publications |
[Note: Highlighted papers indicate first authorship; (*) indicates equal contribution].
|
Understanding Protein-DNA Interactions by Paying
Attention to Protein and Genomics Foundation
Models
Dhruva Abhijit Rajwade, Erica Wang, Aryan Satpathy, Alex Brace, Hongyu Guo, Arvind Ramanathan, Shengchao Liu, Anima Anandkumar NeurIPS 2024 Foundation Models For Science, AI for New Drug Modalities, Machine Learning in Structural Biology workshops, 2024 Paper / Code Cross-Attention coupled with Protein and Genomics Foundation models to understand Protein-DNA interactions speeds up inference and achieves State-of-the-art performance in predicting contacts in Protein-DNA complexes (using purely sequence data for inference). |
|
Towards Backdoor Mitigation and Adversarial Robustness
in SSL
Dhruva Abhijit Rajwade*, Aryan Satpathy*, Nilaksh* ICASSP 2025 Paper / Code An intuitively elegant and simple defense strategy to defend against standard SSL augmentation invariant frequency based backdoor attacks. Taking a leaf out of frequency domain attacks, we also use frequency domain patching to increase model robustness in SSL. |
|
Attenuated Total Reflectance–Fourier Transform Infrared
(ATR-FTIR) Spectroscopy Combined With Deep Learning for
Classification of Idiopathic Recurrent Spontaneous
Miscarriage (IRSM)
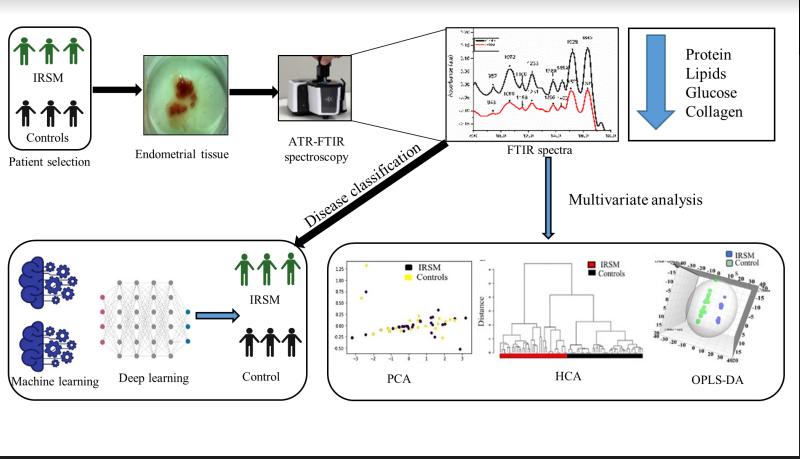
Dadoma Sherpa, Dhruva Abhijit Rajwade, Imon Mitra, Souvik Biswas, Sunita Sharma, Pratip Chakraborty, Shovandeb Kalapahar, Ratna Chattopadhyay, Koel Chaudhury Analytical Letters (Journal), 2024 Paper / Code An extension of our previous work (see below) on using (ATR-FTIR)Spectroscopy and Deep Learning for prediction of Idiopathic Recurrent Spontaneous Miscarriage (IRSM). This work focuses on the classification of IRSM using ATR-FTIR Spectroscopy, which is a non-invasive and cost-effective technique and improves on our previous work on using Raman Spectroscopy in a similar problem setting. |

|
Cells2Vec: Bridging the gap between experiments and
simulations using causal representation learning
Dhruva Abhijit Rajwade, Atiyeh Ahmadi, Brian Ingalls NeurIPS 2023 Causal Representation Learning Workshop, 2023 Paper / Poster / Code / Slides / Talk Learning meaningful representations of multi-cell timeseries (Cellmodeller) simulations using causal representation learning. Current work includes extending this to real-world data, and for proxy-simulation generation for biological experiments. |
|
Prediction of Idiopathic Recurrent Spontaneous
Miscarriage using Machine Learning
[Best Paper Award]
Dadoma Sherpa, Dhruva Abhijit Rajwade, Imon Mitra, Dhruba Dhar, Sunita Sharma, Pratip Chakraborty, Koel Chaudhury IEEE International Conference on Computer, Electrical & Communication Engineering, 2023 Paper / Code Using Raman Spectroscopy and Machine Learning for prediction of Idiopathic Recurrent Spontaneous Miscarriage (IRSM). Improved this work using ATR-FTIR Spectroscopy in a follow-up study, and currently working on a multi-omics approach to the same problem. |
|
Risk factors associated with mortality in
hypersensitivity pneumonitis: a meta-analysis
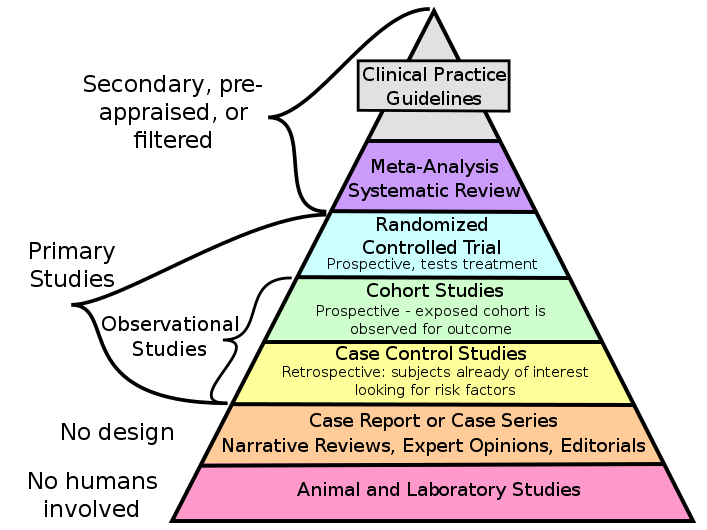
Sanjukta Dasgupta, Anandita Bhattacharya, Dhruva Abhijit Rajwade, Sushmita Roy Chowdhury, Koel Chaudhury Expert Review of Respiratory Medicine (Journal), 2022 Paper / Code Using different statistical tests and empirical analyses to identify risk factors associated with mortality in Hypersensitivity pneumonitis, a rare lung disease. Checking for Publication bias and heterogeneity in the data, and using meta-analysis to combine results from different studies. |
Miscellaneous |
|
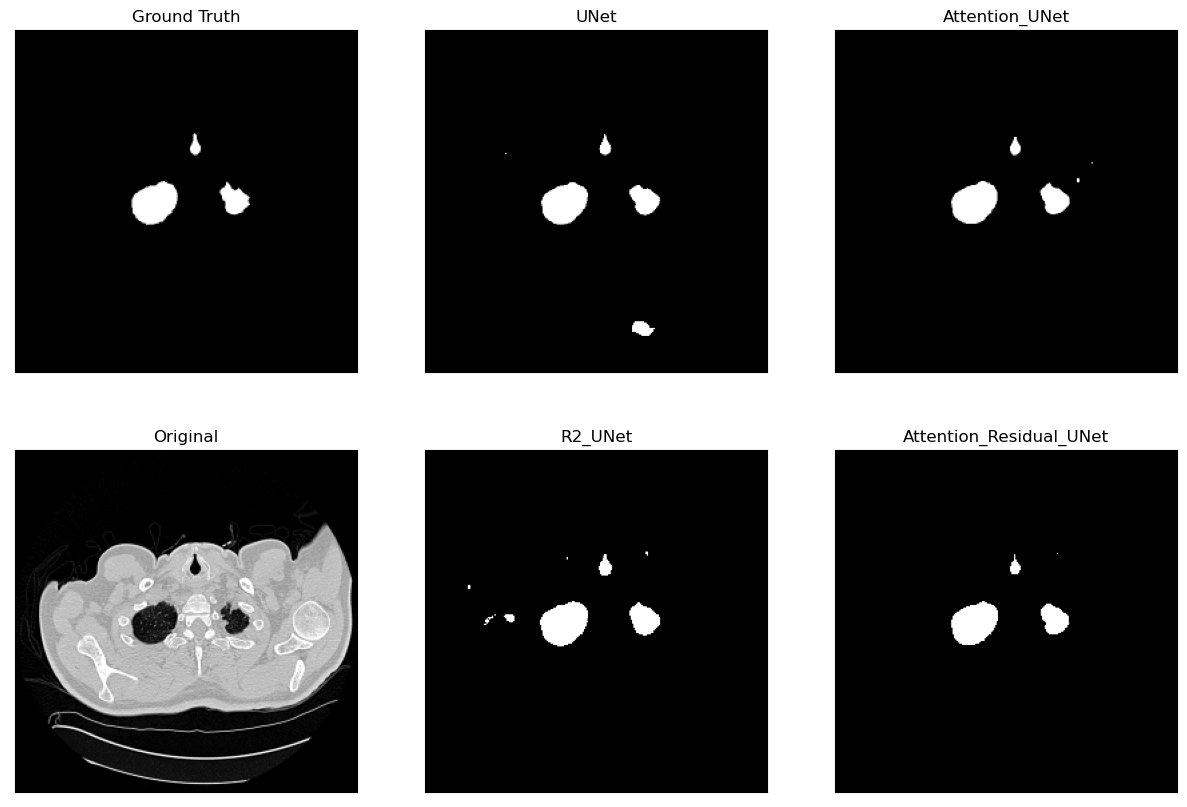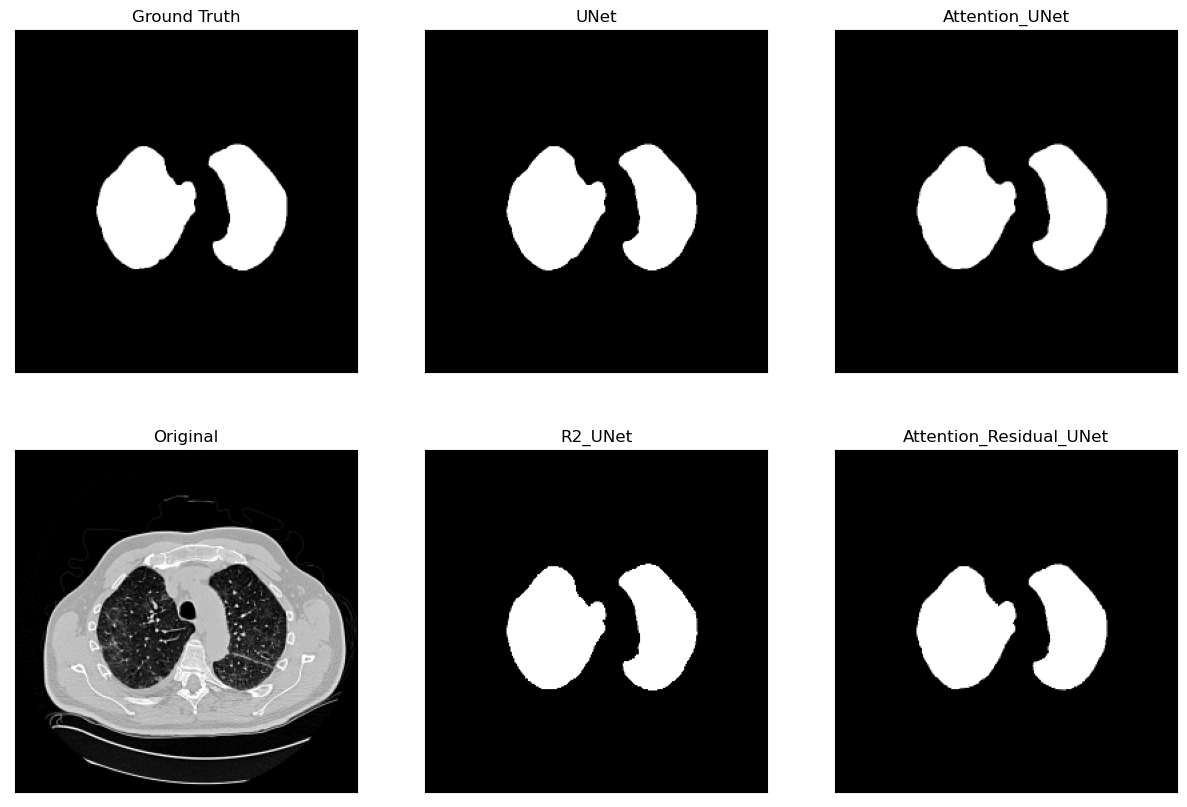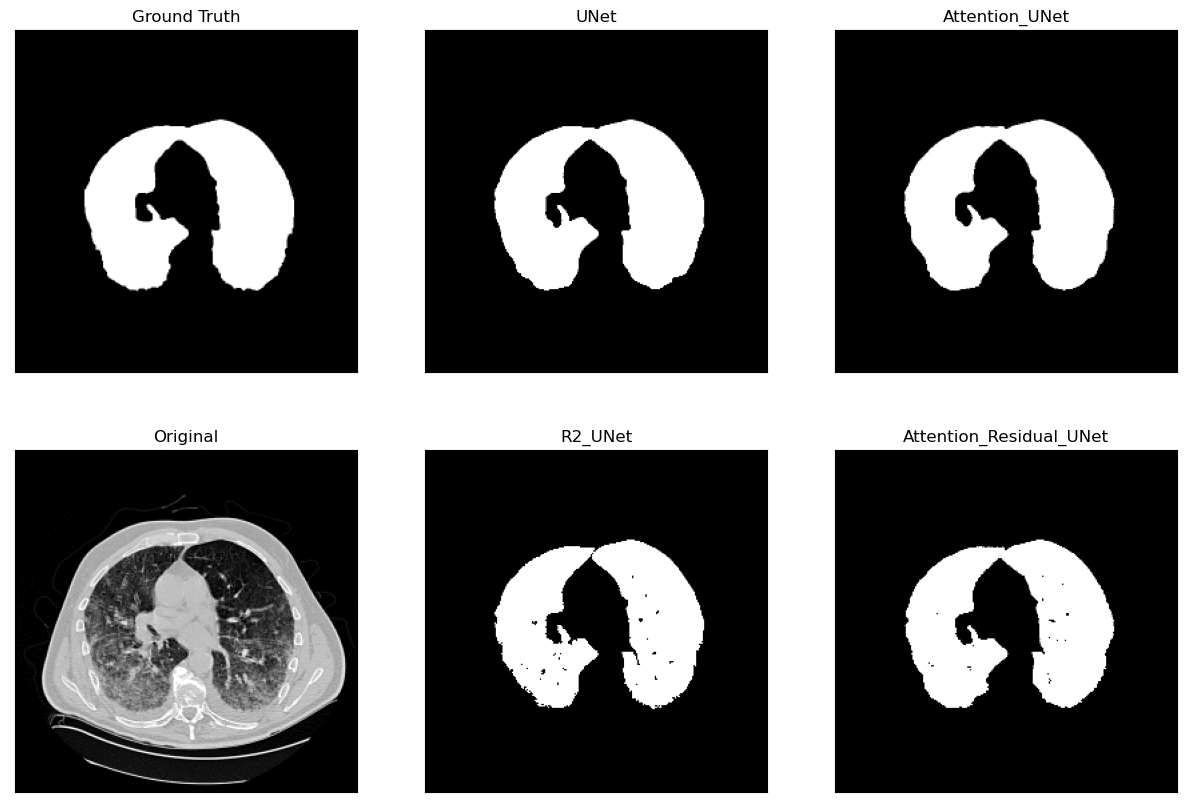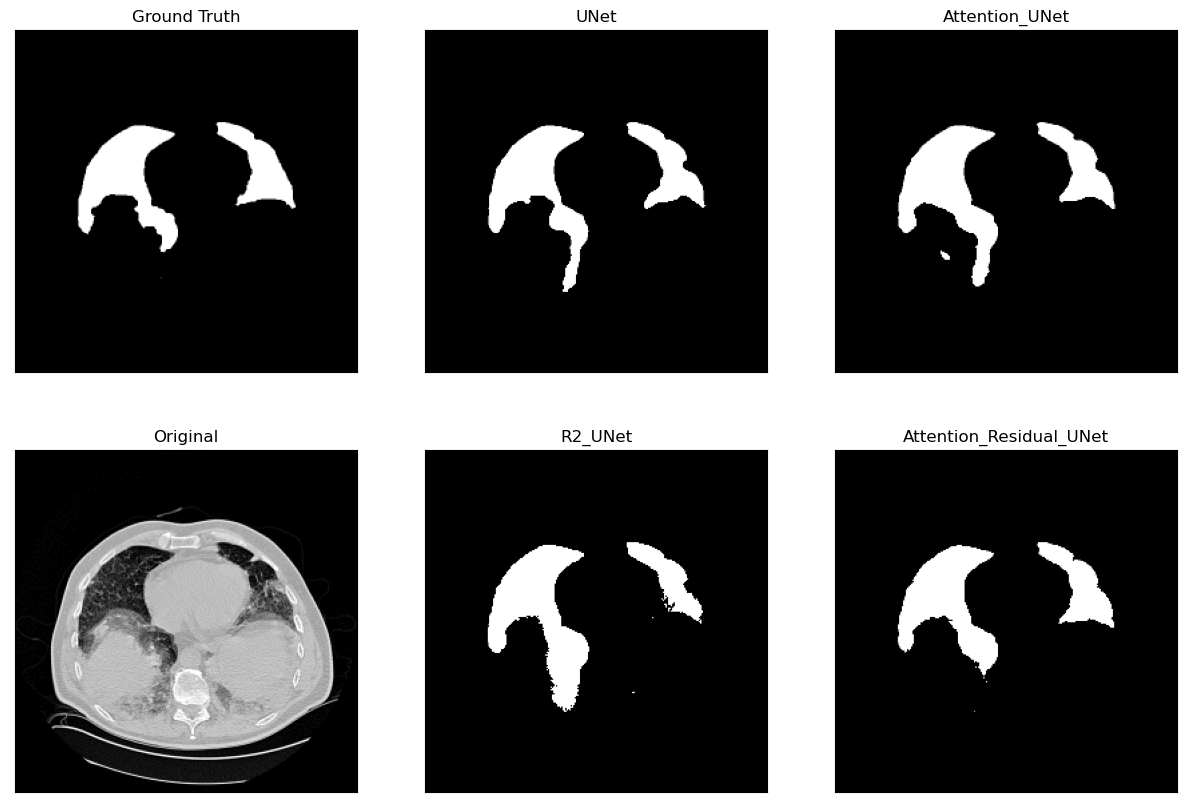
Lung Segmentation and Disease Classification using Deep
Learning
[Code] Worked on using the Geneva HRCT dataset to segment lungs and classify diseases using Deep Learning. The final model uses a U-Net architecture for segmentation and a CNN for classification. The model was trained on a subset of the dataset and tested on CT scan images obtained externally through collaborations with Hospitals. |
|
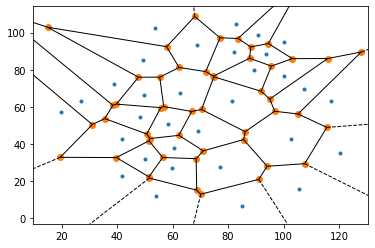
Functional Network Analysis of Calcium ion pathways in
beta-islets of the Pancreas
[Code] Worked on using Calcium ion imaging time-series data to extract functional networks in beta-islets of the pancreas. Used Voronoi Delaunay triangulation (see image) to extract the network, and used graph theory to analyze the network. Code is incomplete and will be updated soon. |
|
This website's template is taken from Jon Barron. Do not scrape the HTML from this page itself, as it includes analytics tags that you do not want on your own website — use the github code instead. Also, consider using Leonid Keselman's Jekyll fork of this page. |